Welcome! I am Fabian Klute, currently I am a PostDoctoral researcher at UPC in Barcelona with the research group on Discrete, Combinatorial and Computational Geometry Geometric Computing. I come from Landau/Palatina in Germany and studied my Bachelor's and Master's degree in Karlsruhe. Recently, In 2020 I finished my PhD at the TU Wien with Martin Nöllenburg.
My main research interests are in the area of graph drawing and automated cartography. At the moment I am very interested in map labeling, content generation, and confluent drawings, as well as parametrized complexity and its application to various geometric topics.
My main research interests are in the area of graph drawing and automated cartography. At the moment I am very interested in map labeling, content generation, and confluent drawings, as well as parametrized complexity and its application to various geometric topics.
- December 2022 – present
- PostDoc in the UPC research group on Discrete, Combinatorial and Computational Geometry at Universitat Politècnica de Catalunya with Prof. Rodrigo Silveira
- April 2020 – December 2022
- PostDoc in the Geometric Computing group at Utrecht University with Prof. Marc J. van Kreveld
- April 2016 – March 2020
- PhD Student in the Algorithms and Complexity Group at TU Wien under the supervision of Prof. Martin Nöllenburg
- May 2015 – December 2015
- Gap months, some impressions can be found here
- April 2009 – April 2015
- Master and Bachelor in Informatics at the Karlsruher Institute of Technology (KIT)
Peer reviewed publications
2022
In Computational Geometry: Theory and Applications, vol. 104, pp. 101860 (2022).
2021
In Journal of Graph Algorithms and Applications, vol. 25, no. 1, pp. 481–512 (2021).
In Algorithmica, vol. 83, no. 1, pp. 297–336 (2021).
In 47th International Conference on Current Trends in Theory and Practice of Computer Science (SOFSEM'21), vol. 12607 of LNCS, pp. 89–103. Springer (2021).
2020
2019
In Journal of Computational Geometry, vol. 10, no. 2, pp. 45–69 (2019).
Extending to 1-plane drawings
In Abstracts of the XVIII Spanish Meeting on Computational Geometry (EGC'19), pp. 30–33.(2019).
In 27th International Symposium on Graph Drawing and Network Visualization (GD'19), vol. 11904 of LNCS, pp. 323–336. Springer (2019).
Theses
Avoiding Crossings in Non-Planar Graph Layouts
TU Wien, 2020
Connecting points with low-complexity polynomial curves in a polygon
Karlsruhe Institute of Technology (KIT), 2015
Serealisierung und Deserialisierung von Mustern in Zellularautomaten
Karlsruhe Institute of Technology (KIT), 2012
GD'18: 4th place
GD'17: 1st place
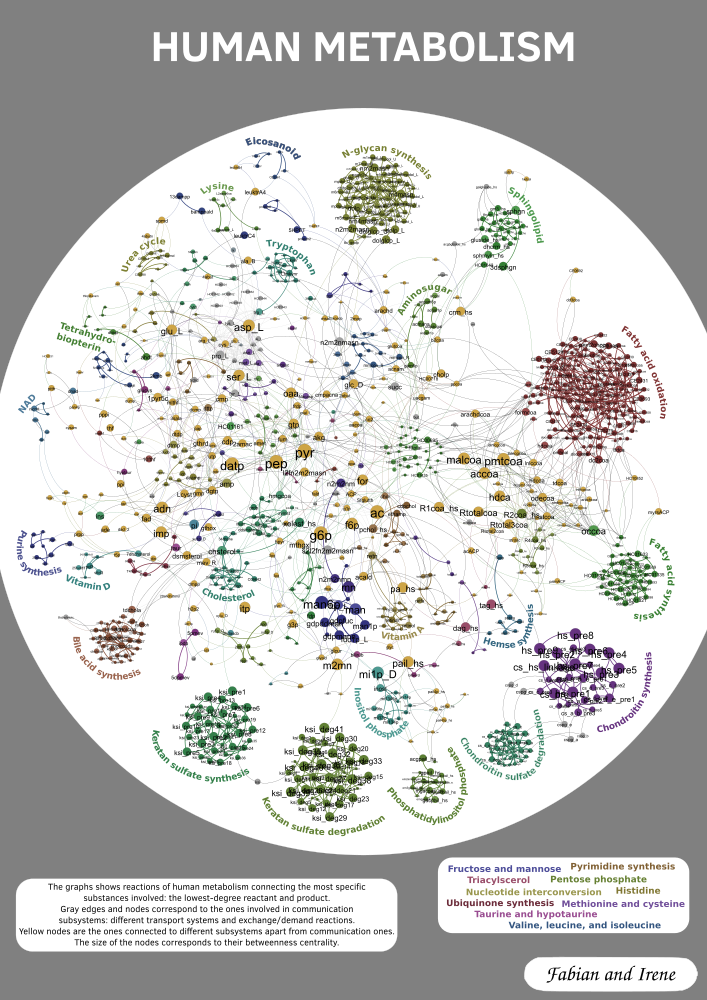
Submission for the GD Contest 2017 together with Irene Parada. The data is taken from the human metabolism project. In order to filter the information, we thin out the graph in a preprocessing step. For every reaction we keep only the lowest-degree reactant and product. Isolated and degree one nodes are removed and only the biggest component is kept. Additionally we identify nodes corresponding to different biological compartments (e.g. co2[c], co2[e] are contracted into one co2 node). To generate the layout we add edges to keep the subsystems closer together. The size of the nodes depends on its betweenes in the reduced graph. Then the layout is calculated using the Fruchterman-Reingold algorithm. Finally we color the nodes based on subsystems. We distinguish two big groups of subsystems, the communication or transportation ones and the proper ones. As the name suggests the first kind mainly connects the proper subsystems. Based on this we categorize nodes into five cases, depending on if they are inside a proper subsystem, on its boundary, in a transport system, on the boundary between two different transport systems, or between two proper subsystems.
GD'16: 1st place
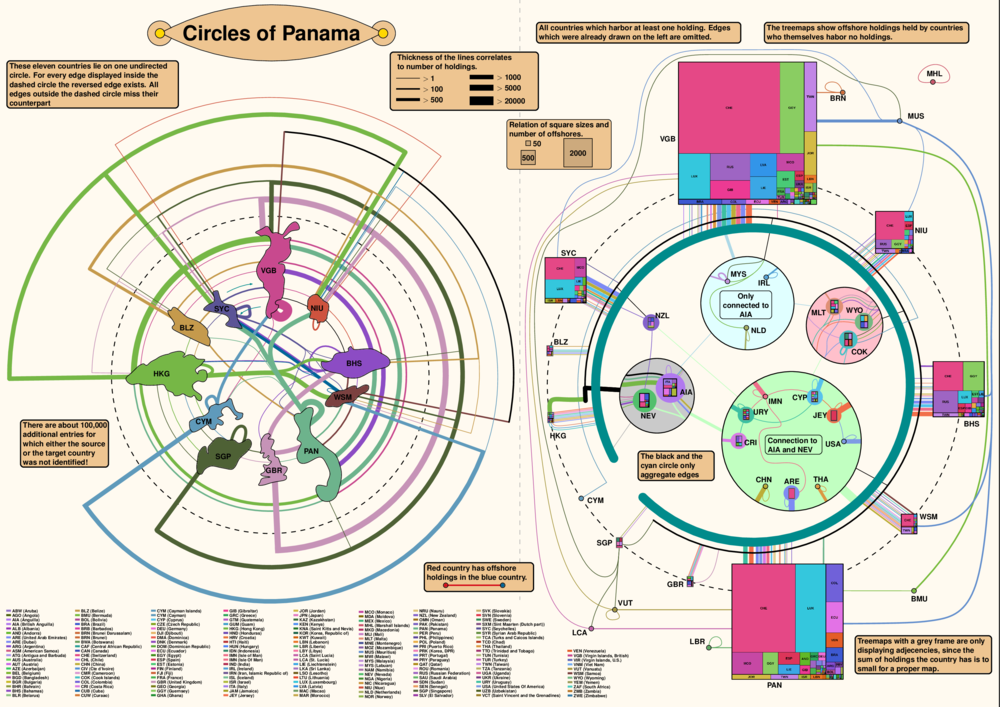
Submission for the GD Contest 2016. The data is an experpt of the data surfaced in the panama papers. Graph and layout were produced by exploring the graph with yed, R, whatever C program I had lying around. I focused mainly on finding sets of strongly connected vertices. I noticed a group of eleven vertices which are connected by an undirected circle and on top are almost all connected to each other in some way. I extracted this group and made one visualization for them in Ipe (left half). Among the leftover vertices a lot of them have no incoming edges so I plotted them in treemaps using R. These treemaps I then used as vertices and connected them with the other remaining vertices with incoming edges that were not part of the above mentioned group (right half). All layouting was done in Ipe by hand with the help of a couple of python scripts generating e.g. the legend.
Find me at Universitat Politècnica de Catalunya
Edifici Omega (office 419), Campus Nord
Jordi Girona, 1-3
08034 Barcelona
Spain